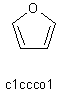SMILES (Simplified Molecular Input Line Entry System) is a line notation (a typographical method using printable characters) for entering and representing molecules and reactions.
SMILES contains the same information as might be found in an extended connection table. The primary reason SMILES is more useful than a connection table is that it is a linguistic construct, rather than a computer data structure. SMILES is a true language, albeit with a simple vocabulary (atom and bond symbols) and only a few grammar rules. SMILES representations of structure can in turn be used as "words" in the vocabulary of other languages designed for storage of chemical information (information about chemicals) and chemical intelligence (information about chemistry).
Part of the power of SMILES is that unique SMILES exist. With standard SMILES, the name of a molecule is synonymous with its structure; with unique SMILES, the name is universal. Anyone in the world who uses unique SMILES to name a molecule will choose the exact same name.
One other important property of SMILES is that it is quite compact compared to most other methods of representing structure. A typical SMILES will take 50% to 70% less space than an equivalent connection table, even binary connection tables. For example, a database of 23,137 structures, with an average of 20 atoms per structure, uses only 1.6 bytes per atom when represented with SMILES. In addition, ordinary compression of SMILES is extremely effective. The same database cited above was reduced to 27% of its original size by Ziv-Lempel compression (i.e. 0.42 bytes per atom).
These properties open many doors to the chemical information programmer. Examples of uses for SMILES are:
- Keys for database access
- Mechanism for researchers to exchange chemical information
- Entry system for chemical data
- Part of languages for artificial intelligence or expert systems in chemistry
Structural images of SMILES
| Structural image | SMILES notation |
| C=CC\C=C\O |
| CCN(CC)CC |
| CC(C)C(=O)O |
| CC(C)C(CCC)C(CCC)C=C |
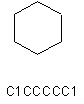
| C1CCCCC1 |
| CC1=CC(Br)CCC1 |
| C1CN(CCC1)C2CCCCO2 |
| c1ccco1 |
| Oc1ccncn1 |
| c1ccccn1 |
| ON1CCCCC1 |
| O[n+]1ccccc1 |
| Oc1ccccc1 |
| Cn1cccc1 |
| C[C@H]=C\C=C\F |
| c1cccn1 |
| Oc1ccccn1 |
| C\C=C\C=C\F |